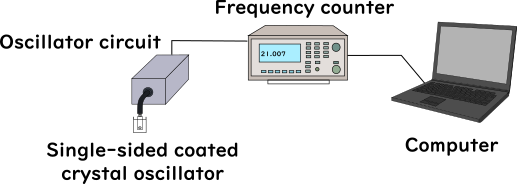
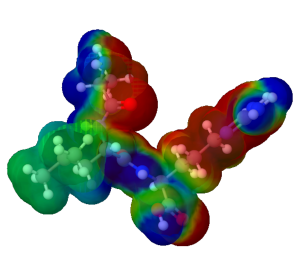
The Iiage of electrostatic potential of tripeptide
—Authentication demo animation—
—Authentication demo animation—


 Fit
Not fit
Fit
Not fit
We started the development of "In sillico software"
We started the new project to search the low molecular weight compounds
( activators or inhibitors ) with "In silico software".
" In silico software " includes to find new pathways
with the setting of the weighting factors.
We plan to make "In silico software" more accurate and reliable
by applying "image recognition" and "deep learning" technology
and our original evaluation systems (in vitro・in situ・in vivo).
We started the new project to search the low molecular weight compounds
( activators or inhibitors ) with "In silico software".
" In silico software " includes to find new pathways
with the setting of the weighting factors.
We plan to make "In silico software" more accurate and reliable
by applying "image recognition" and "deep learning" technology
and our original evaluation systems (in vitro・in situ・in vivo).
Maximizing efficiency for searching for target compounds
using【 machine (reinforcement)learning 】
using【 machine (reinforcement)learning 】

Candidate compounds
Cyclophilin
ProteinーCompound Docking Learning
Examination of Auto Dock /
Empirical Score ( PRO-LEAD,
GOLD chemscore etc. )

Image recognation
programming

Protein structure prediction
and modeling
Narrowing down of target compounds

Confirmation of intermolecular interaction using
QCM ( Quarts Crystal Microbalance ) bio-sensor